Z4 - Bioinformatic support for the CRC
During the 3rd funding period we will provide bioinformatics support for the other projects. This encompasses advising, development and application of analysis strategies, which will then contiguously be implemented as Galaxy workflows. The Galaxy instance, as well as the workflows will be maintained, updated and adapted to stay up-to date and usable. We will consistently train members of the CRC in using the Galaxy system. The integration of multi-omics data using network approaches will be the methodical focus.
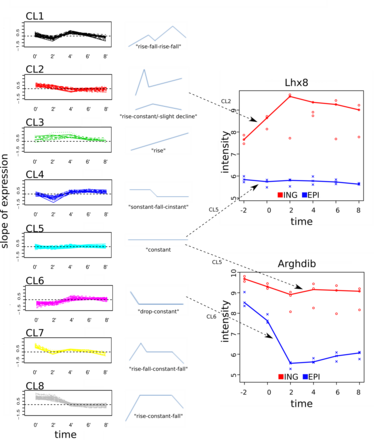
Figure 1: Novel cluster approach to compare gene expression time course in distinct adipose tissues. A) regardless of fat depot (ING: inguinal, EPI: epididymal) the slopes of gene expression profiles are clustered (left panel). Each cluster represents a general gene expression trend over time (middle panel). The combination of cluster assignments to each fat depot was used to retrieve genes with different dynamics over time (-2: baseline expression, 0: differentiation stimulus, 2 days of adipogenesis (doa), 4 doa, 6 doa ,8 doa). Right panel: Examples with approx. constant expression (cluster 5 (CL5)) in adipocytes in one of the fat depots, while Lhx8 expression initially rises (cluster 2 (CL2)), and Arghdib initially drops (cluster 6 (CL6)) in the cells of the respective other fat depot.
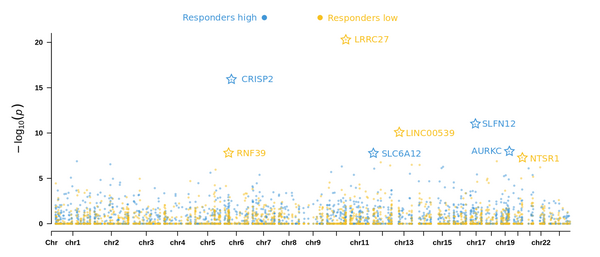
Figure 2: Epigenome wide association study for weight loss identifies genes significantly correlated with weight loss in high responders (blue) and low responders (yellow).
Krieg L, Didt K, Karkossa I, Bernhart SH, Kehr S, Subramanian N, Lindhorst A, Schaudinn A, Tabei S, Keller M, Stumvoll M, Dietrich A, von Bergen M, Stadler PF, Laurencikiene J, Krüger M, Blüher M, Gericke M, Schubert K, Kovacs P, Chakaroun R, Massier L. Gut. 2021 Oct 1:gutjnl-2021-324603.
Kolora SRR, Gysi DM, Schaffer S, Grimm-Seyfarth A, Szabolcs M, Faria R, Henle K, Stadler PF, Schlegel M, Nowick K. Accelerated evolution of tissue-specific genes mediates divergence amidst gene flow in European green lizards. Genome Biol Evol. 2021 May 14:evab109.
Yaskolka Meir A, Keller M, Müller L, Bernhart SH, Tsaban G, Zelicha H, Rinott E, Kaplan A, Gepner Y, Shelef I, Schwarzfuchs D, Ceglarek U, Stadler PF, Blüher M, Stumvoll M, Kovacs P, Shai I. Effects of lifestyle interventions on epigenetic signatures of liver fat: CENTRAL randomized controlled trial. Liver Int. 2021 May 3.
Meir A Y, Keller Maria, Bernhart S H, Rinott E, Tsaban G, Zelicha H, Kaplan A, Schwarzfuchs D, Shelef I, Gepner Y, Li J, Lin Y, Blüher M, Ceglarek U, Stumvoll M, Stadler PF, Stampfer M J, Kovacs P, Liang L, Shai I. Lifestyle weight‑loss intervention may attenuate methylation aging: the CENTRAL MRI randomized controlled trial. Clin Epigenetics. 2021 Mar 4;13(1):48.
Lorenz R, Stadler PF. RNA Secondary Structures with Limited Base Pair Span: Exact Backtracking and an Application. Genes (Basel). 2020 Dec 24;12(1):E14.
Hoser SM, Hoffmann A, Meindl A, Gamper M, Fallmann J, Bernhart SH, Müller L, Ploner M, Misslinger M, Kremser L, Lindner H, Geley S, Schaal H, Stadler PF, Huettenhofer A. Intronic tRNAs of mitochondrial origin regulate constitutive and alternative splicing. Genome Biol. 2020 Dec 8;21(1):299.
Keller M, Yaskolka Meir A, Bernhart SH, Gepner Y, Shelef I, Schwarzfuchs D, Tsaban G, Zelicha H, Hopp L, Müller L, Rohde K, Böttcher Y, Stadler PF, Stumvoll M, Blüher M, Kovacs P, Shai I. DNA methylation signature in blood mirrors successful weight-loss during lifestyle interventions: the CENTRAL trial. Genome Med. 2020 Nov 16;12(1):97.
Nono Nankam PA, Blüher M, Kehr S, Klöting N, Krohn K, Adams K, Stadler PF, Mendham AE, Goedecke JH. Distinct abdominal and gluteal adipose tissue transcriptome signatures are altered by exercise training in African women with obesity. Sci Rep. 2020 Jun 24;10(1):10240.
Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 2016;529:496-501.
Jühling F, Kretzmer H, Bernhart SH, Otto C, Stadler PF, Hoffmann S. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;6:256-62.
Liu X, Hinney A, Scholz M, Scherag A, Tönjes A, Stumvoll M, Stadler PF, Hebebrand J, Böttcher Y. Indications for potential parent-of-origin effects within the FTO gene. PLoS One. 2015;10:e0119206.
Hoffmann S, Stadler PF, Strimmer K. A simple data-adaptive probabilistic variant calling model. Algorithms Mol Biol. 2015;10:10.
Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt LM, Teupser D, Hackermüller J, Stadler PF. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34.
Otto C, Stadler PF, Hoffmann S. Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics. 2014;30:1837-43.
Hackermüller J, Reiche K, Otto C, Hösler N, Blumert C, Brocke-Heidrich K, Böhlig L, Nitsche A, Kasack K, Ahnert P, Krupp W, Engeland K, Stadler PF, Horn F. Cell cycle, oncogenic and tumor suppressor pathways regulate numerous long and macro non-protein-coding RNAs. Genome Biol. 2014;5:R48.
Nitsche A, Doose G, Tafer H, Robinson M, Saha NR, Gerdol M, Canapa A, Hoffmann S, Amemiya CT, Stadler PF. Atypical RNAs in the coelacanth transcriptome. J Exp Zool B Mol Dev Evol. 2014;322:342-51.
Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, Finstermeier K, Stahringer A, Wilfert W, Beutner F, Gielen S, Schuler G, Gäbel G, Bergert H, Bechmann I, Stadler PF, Thiery J, Teupser D. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013;9:e1003588.
Otto C, Stadler PF, Hoffmann S. Fast and sensitive mapping of bisulfite-treated sequencing data. Bioinformatics. 2012;28:1698-704.